Different viruses, including influenza A and HIV, mutate so rapidly that developing highly effective therapies and vaccines are like trying to hit an itinerant target. Improved understanding of viral transmission and development in single cells could help. At present, researchers report a new method to identify and quantify viral RNA in living cells that also aids to detect slight changes in RNA sequences that might give viruses an edge or make some people “superspreaders.”
The researchers offered their findings at the American Chemical Society (ACS) Fall 2020 Virtual Meeting & Expo. It featured innovative and emerging more than 6000 presentations under the wide spectrum of science topics.
“Studies for a new virus like SARS-CoV-2, it’s important to learn the response of not only populations but individuals – either people or cells – interacting with it,” says Laura Fabris, Ph.D., the project’s principal investigator. “So we’ve focused on learning viral replication in single cells, which in the past has been theoretically challenging.”
Instead of large populations analyzing individual cells could go a long way toward an enhanced understanding of many facets of viral outbreaks like superspreaders. In this phenomenon, certain individual cells or people strangely carry the virus in high amounts causing to infect many others. If scientists could identify individual cells with elevated viral loads in superspreaders and then study the viral sequences in those cells, they could find out how viruses evolve to become more transmittable or to outwit treatments and vaccines. Furthermore, the characteristics of the host cell itself could help many viral progressions and consequently become targets for therapies. On the other end of the spectrum, some cells make mutated viruses that are no longer infectious. Understanding how this takes place could also lead to new and effective vaccines and antiviral therapies.
Fabris and colleagues at Rutgers University are firstly required to build up an assay that was sensitive enough to sense viral RNA, and its mutations, in single living cells. The researchers based their technique on surface-enhanced Raman spectroscopy (SERS), an insightful method that detects connections between molecules through changes in how they disperse light. To study influenza A the researcher’s team decided to use this method. To detect the virus’s RNA, they added to gold nanoparticles a “beacon DNA” specific to influenza A. In the incidence of influenza A RNA, the beacon produced a well-built SERS signal, while in the absence of this RNA, it did not. The beacon produced less strong SERS signals with increasing numbers of viral mutations, permitting the researchers to detect as few as two nucleotide changes. Significantly, the nanoparticles could enter human cells in a dish, and they produced a SERS signal only in those cells exhibiting influenza A RNA.
Now, Fabris and colleague researchers are making a version of the assay that upon viral RNA is detection produces a fluorescent signal, instead of a SERS signal. But SERS is not a clinically approved technique. It’s just now contravention into the clinic,” Fabris notes. “So we required to present clinicians and virologists with a technology they would be more familiar with and have the technology to use right now.” In collaboration with mathematicians and virologists at other universities, the researchers are developing microfluidic procedure, or “lab-on-a-chip” technologies, to read many fluorescent samples at the same time.
Because SERS is cheaper, faster, sensitive and convenient to perform than other assays based on fluorescence or the reverse transcriptase-polymerase chain reaction (known as RT-PCR), it could prove perfect for detecting and examining viruses in the future. Fabris is now actively collaborating with a company that makes cheap, portable Raman spectrometer, which would facilitate the SERS assay to be easily conducted in the field.
Fabris and her team are also working on identifying regions of the SARS-CoV-2 genome to target with SERS probes. “We’re in the developmental stages of obtaining financial assistance to work on possible SARS-CoV-2 diagnostics with the SERS method we developed,” Fabris says.
The researchers acknowledge funding and technical support from the Defense Advanced Research Projects Agency.

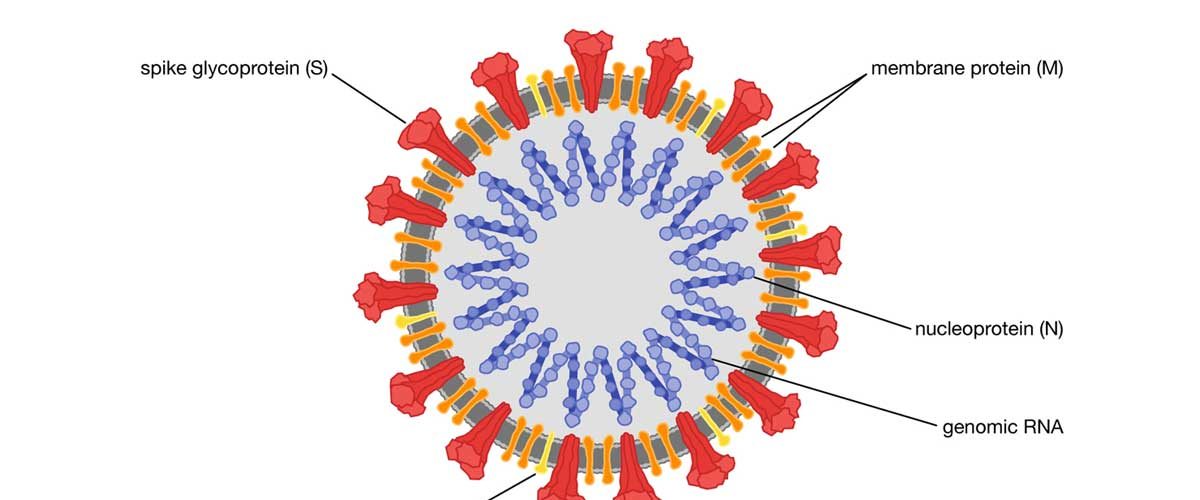










Very helpful study to develop a potential vaccine or treatment against COVID-19
In my opinion, the selection of systemic experimental and computational methods influence greatly the research findings.